Covariate adjustment in randomized experiments
The post motivated by a tweetorial from Darren Dahly
In an experiment, do we adjust for covariates that differ between treatment levels measured pre-experiment (“imbalance” in random assignment), where a difference is inferred from a t-test with p < 0.05? Or do we adjust for all covariates, regardless of differences pre-test? Or do we adjust only for covariates that have sustantial correlation with the outcome? Or do we not adjust at all?
The original tweet focussed on Randomized Clinical Trials, which typically have large sample size. Here I simulate experimental biology, which typically has much smaller n.
library(ggplot2)
library(GGally)
library(data.table)
source("../R/fake_x.R") # bookdownFake data
Generate \(p\) correlated variables and assign the first to the response (\(\mathbf{y}\)) and the rest to the covariates (\(\mathbf{X}\)). Construct a treatment variable and effect and add this to the response.
n <- 100 # per treatment level - this is modified below
p <- 3 # number of covariates (columns of the data)
pp1 <- p+1
beta_0 <- 0 # intercept
niter <- 2000 # modified below
measure_cols <- c("no_adjust", "imbalance", "all_covariates", "weak_covariates", "strong_covariates")
xcols <- paste0("X", 1:p)
build_ycols <- c("Y_o", xcols)
cor_ycols <- c("Y", xcols)
b_mat <- data.table(NULL)
se_mat <- data.table(NULL)
p_mat <- data.table(NULL)
ci_mat <- data.table(NULL)
for(beta_1 in c(0, 0.2, 0.8)){ # treatment effect on standardized scale
beta <- c(beta_0, beta_1)
for(n in c(6, 10, 50)){
# larger iterations with smaller n
niter <- round((3*10^4)/sqrt(n), 0)
niter <- 2000
# repopulate with NA each n
b <- se <- pval <- ci <- matrix(NA, nrow=niter, ncol=length(measure_cols))
colnames(b) <- colnames(se) <- colnames(pval) <- colnames(ci) <- measure_cols
Treatment <- rep(c("Cn", "Tr"), each=n)
X <- model.matrix(formula("~ Treatment"))
for(iter in 1:niter){
# generate p random, correlated variables. The first is assigned to Y
fake_data <- fake.X(n*2, pp1, fake.eigenvectors(pp1), fake.eigenvalues(pp1))
colnames(fake_data) <- build_ycols
# resacale so that var(Y) = 1, where Y is the first column
fake_data <- fake_data/sd(fake_data[,1])
fake_data <- data.table(fake_data)
# view the scatterplots
#gg <- ggpairs(X,progress = ggmatrix_progress(clear = FALSE))
show_it <- FALSE
if(show_it ==TRUE){
gg <- ggpairs(fake_data)
print(gg, progress = F)
}
# add the treatment effect
fake_data[, Y:=Y_o + X%*%beta]
fake_data[, Treatment:=Treatment]
# model 1 - just the treatment
fit1 <- lm(Y ~ Treatment, data=fake_data)
res <- coef(summary(fit1))["TreatmentTr", ]
b[iter, 1] <- res["Estimate"]
se[iter, 1] <- res["Std. Error"]
pval[iter, 1] <-res["Pr(>|t|)"]
ci_i <- confint(fit1)["TreatmentTr",]
ci[iter, 1] <- ifelse(beta_1 >= ci_i[1] & beta_1 <= ci_i[2], 1, 0)
res1 <- copy(res)
# model 2 - adjust for imablance
inc_xcols <- NULL
for(i in 1:p){
formula <- paste0(xcols[i], " ~ Treatment")
fit2a <- lm(formula, data=fake_data)
if(coef(summary(fit2a))["TreatmentTr", "Pr(>|t|)"] < 0.05){
inc_xcols <- c(inc_xcols, xcols[i])
}
}
if(length(inc_xcols) > 0){ # if any signifianct effects refit, otherwise use old fit
formula <- paste0("Y ~ Treatment + ", paste(inc_xcols, collapse=" + "))
fit2b <- lm(formula, data=fake_data)
res <- coef(summary(fit2b))["TreatmentTr", ]
ci_i <- confint(fit2b)["TreatmentTr",]
}else{
res <- res1
}
b[iter, 2] <- res["Estimate"]
se[iter, 2] <- res["Std. Error"]
pval[iter, 2] <-res["Pr(>|t|)"]
ci[iter, 2] <- ifelse(beta_1 >= ci_i[1] & beta_1 <= ci_i[2], 1, 0)
# model 3- adjust for covariates
(ycor <- abs(cor(fake_data[, .SD, .SDcols=cor_ycols])[2:pp1, 1]))
mean(ycor)
j <- 2
for(target_cor in c(0, .2, .4)){
j <- j+1
if(target_cor == 0.2){
inc <- which(ycor < target_cor) # include only weak covariates
}else{
inc <- which(ycor > target_cor) # include all OR strong covariates
}
if(length(inc) > 0){ # if matches refit, otherwise use old fit
inc_xcols <- xcols[inc]
formula <- paste0("Y ~ Treatment + ", paste(inc_xcols, collapse=" + "))
fit3 <- lm(formula, data=fake_data)
res <- coef(summary(fit3))["TreatmentTr", ]
ci_i <- confint(fit3)["TreatmentTr",]
}else{
res <- res1
}
b[iter, j] <- res["Estimate"]
se[iter, j] <- res["Std. Error"]
pval[iter, j] <-res["Pr(>|t|)"]
ci[iter, j] <- ifelse(beta_1 >= ci_i[1] & beta_1 <= ci_i[2], 1, 0)
}
}
b_mat <- rbind(b_mat, data.table(n=n, beta_1=beta_1, b))
se_mat <- rbind(se_mat, data.table(n=n, beta_1=beta_1, se))
p_mat <- rbind(p_mat, data.table(n=n, beta_1=beta_1, pval))
ci_mat <- rbind(ci_mat, data.table(n=n, beta_1=beta_1, ci))
}
}
p_long <- melt(p_mat, measure.vars=measure_cols, variable.name="method", value.name="p")
ci_long <- melt(ci_mat, measure.vars=measure_cols, variable.name="method", value.name="covers")
b_long <- melt(b_mat, measure.vars=measure_cols, variable.name="method", value.name="b")
se_long <- melt(se_mat, measure.vars=measure_cols, variable.name="method", value.name="se")
#ci_long[, .(coverage=sum(covers)/niter), by=.(method, n, beta_1)]Distribution of estimates
pd <- position_dodge(0.8)
gg <- ggplot(data=b_long, aes(x=factor(n), y=b, fill=method)) +
geom_boxplot(position=pd) +
xlab("sample size (per treatment level)") +
NULL
gg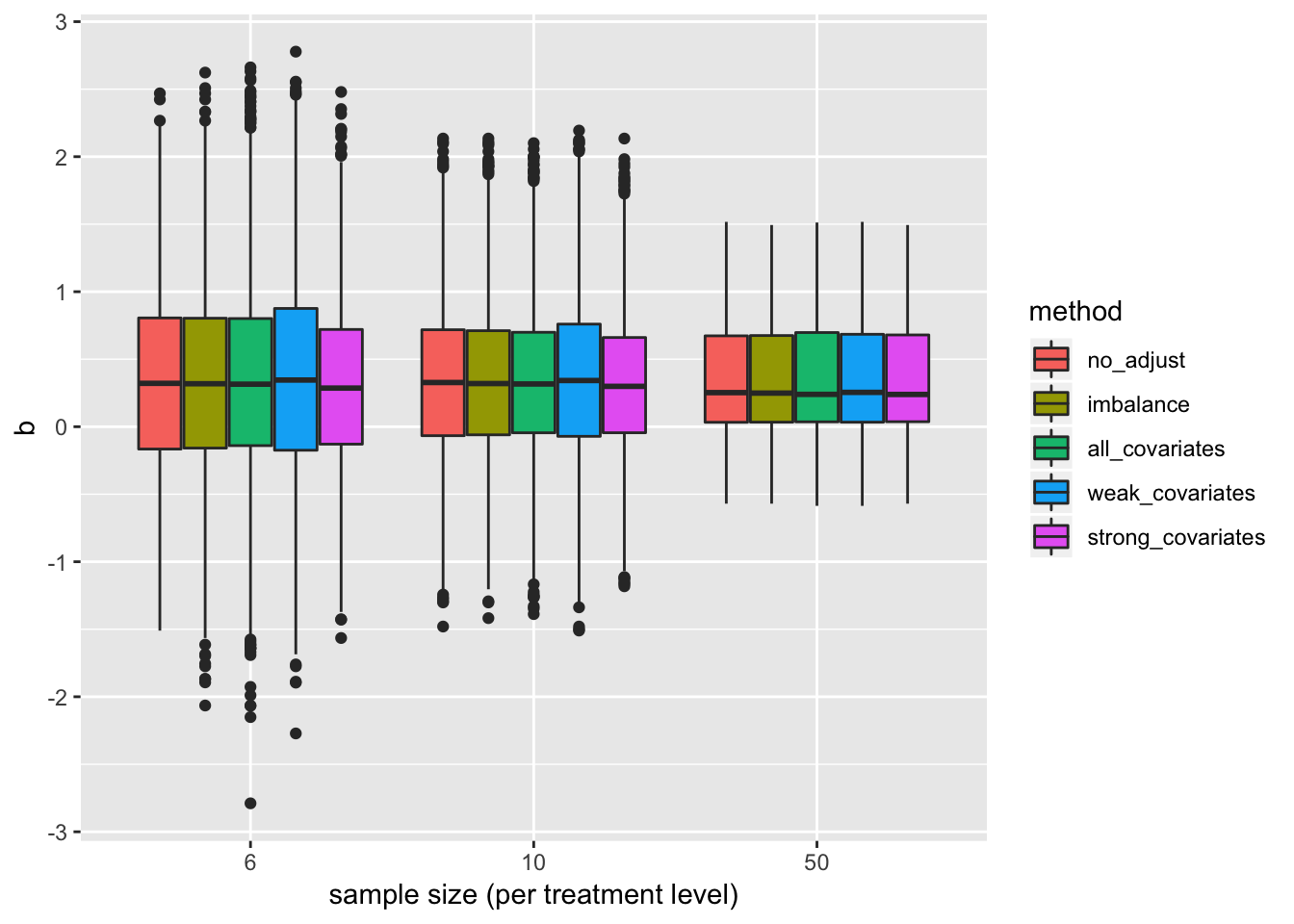
Distribution of SE of estimate
pd <- position_dodge(0.8)
gg <- ggplot(data=se_long, aes(x=factor(n), y=se, fill=method)) +
geom_boxplot(position=pd) +
xlab("sample size (per treatment level)") +
NULL
gg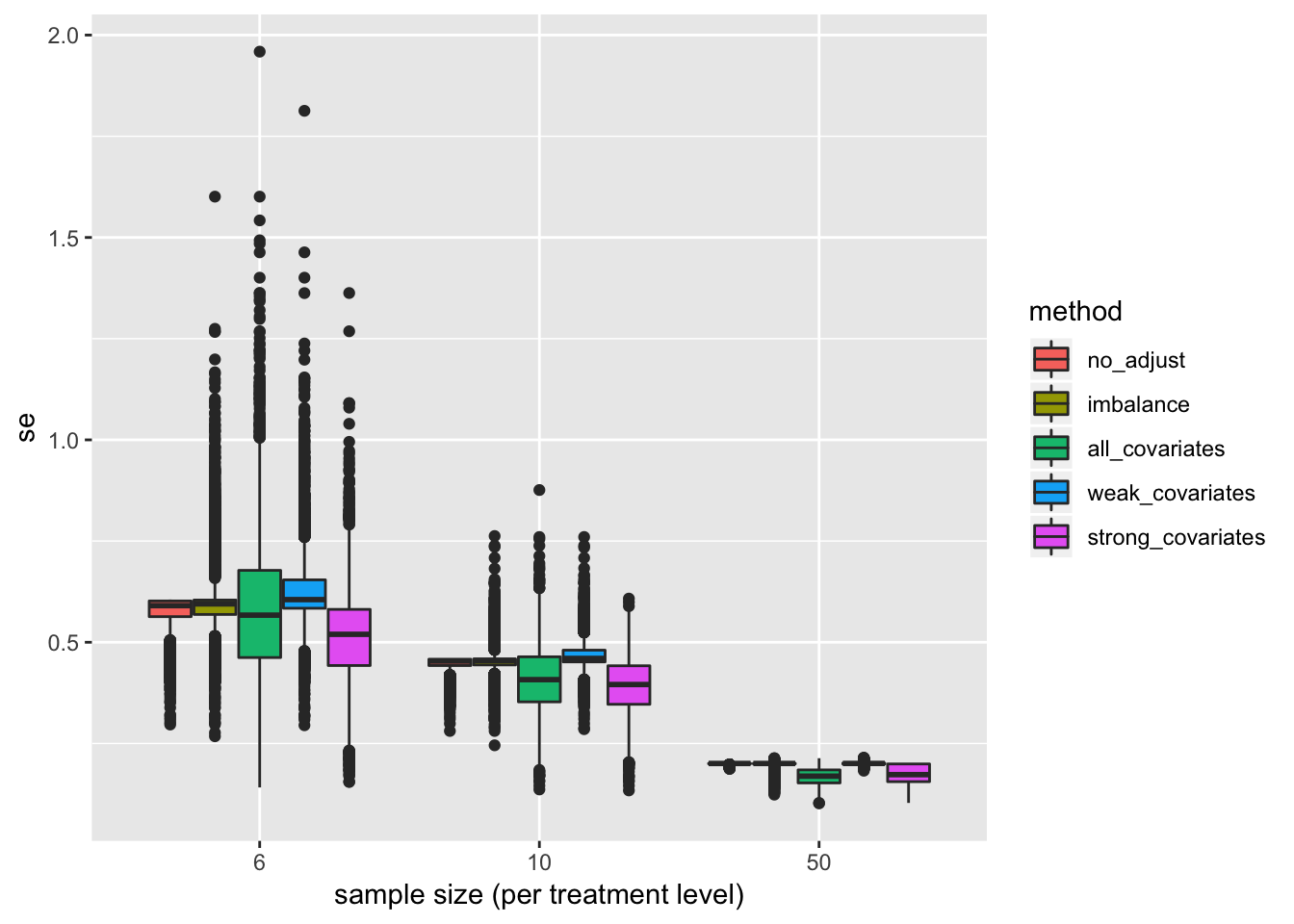
Type I error
# type I
p_sum <- p_long[, .(error=sum(p < 0.05)/niter), by=.(method, n, beta_1)]
pd <- position_dodge(0.8)
gg <- ggplot(data=p_sum[beta_1==0], aes(x=factor(n), y=error, color=method, group=method)) +
geom_point(position=pd) +
geom_line(position=pd) +
xlab("sample size (per treatment level)") +
ylab("Type I error") +
# facet_grid(.~beta_1) +
NULL
gg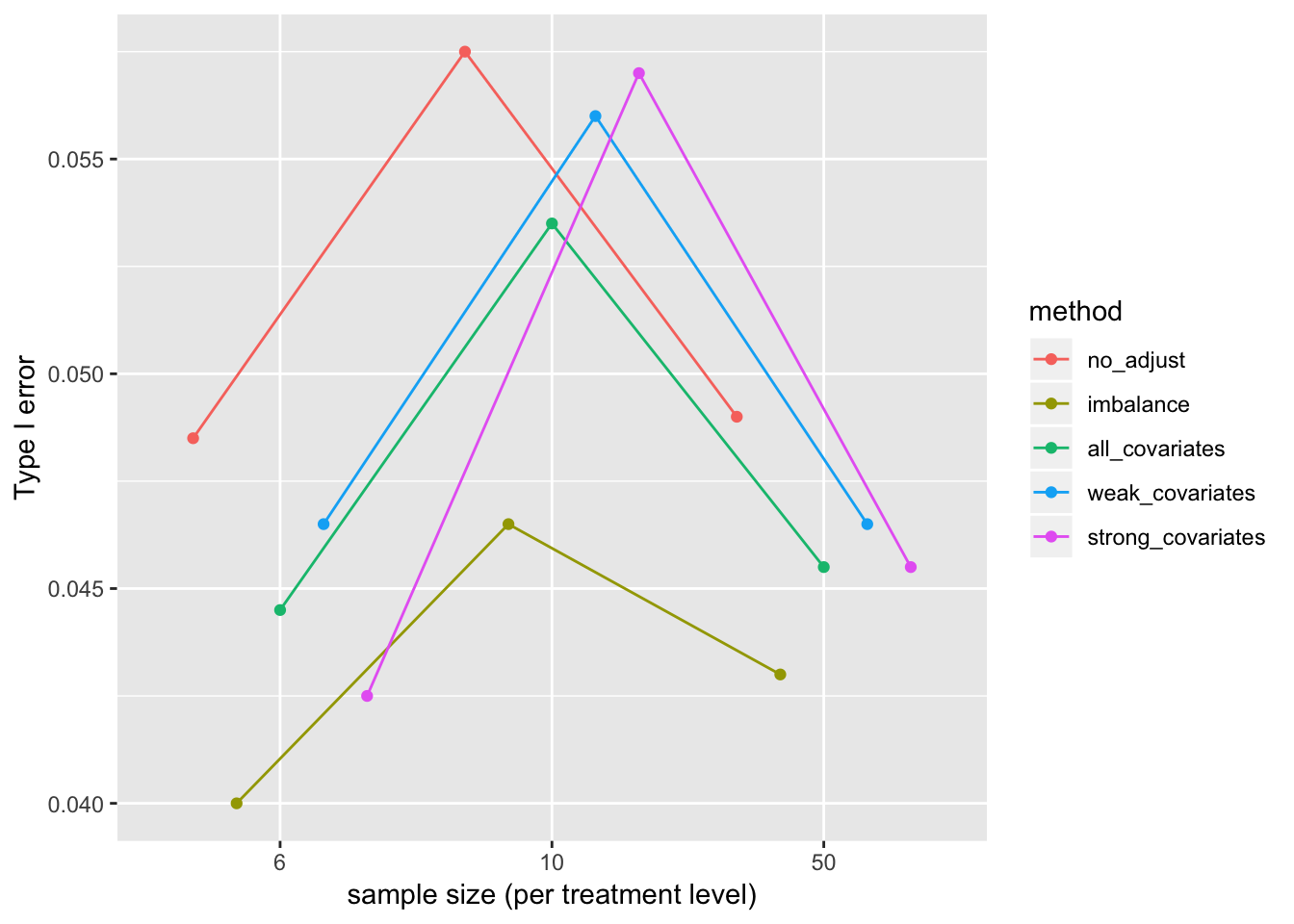
Power
# power
# need p_sum from above
gg <- ggplot(data=p_sum[beta_1!=0], aes(x=factor(n), y=error, color=method)) +
geom_point(position=pd) +
xlab("sample size (per treatment level)") +
ylab("Power") +
facet_grid(.~beta_1) +
NULL
gg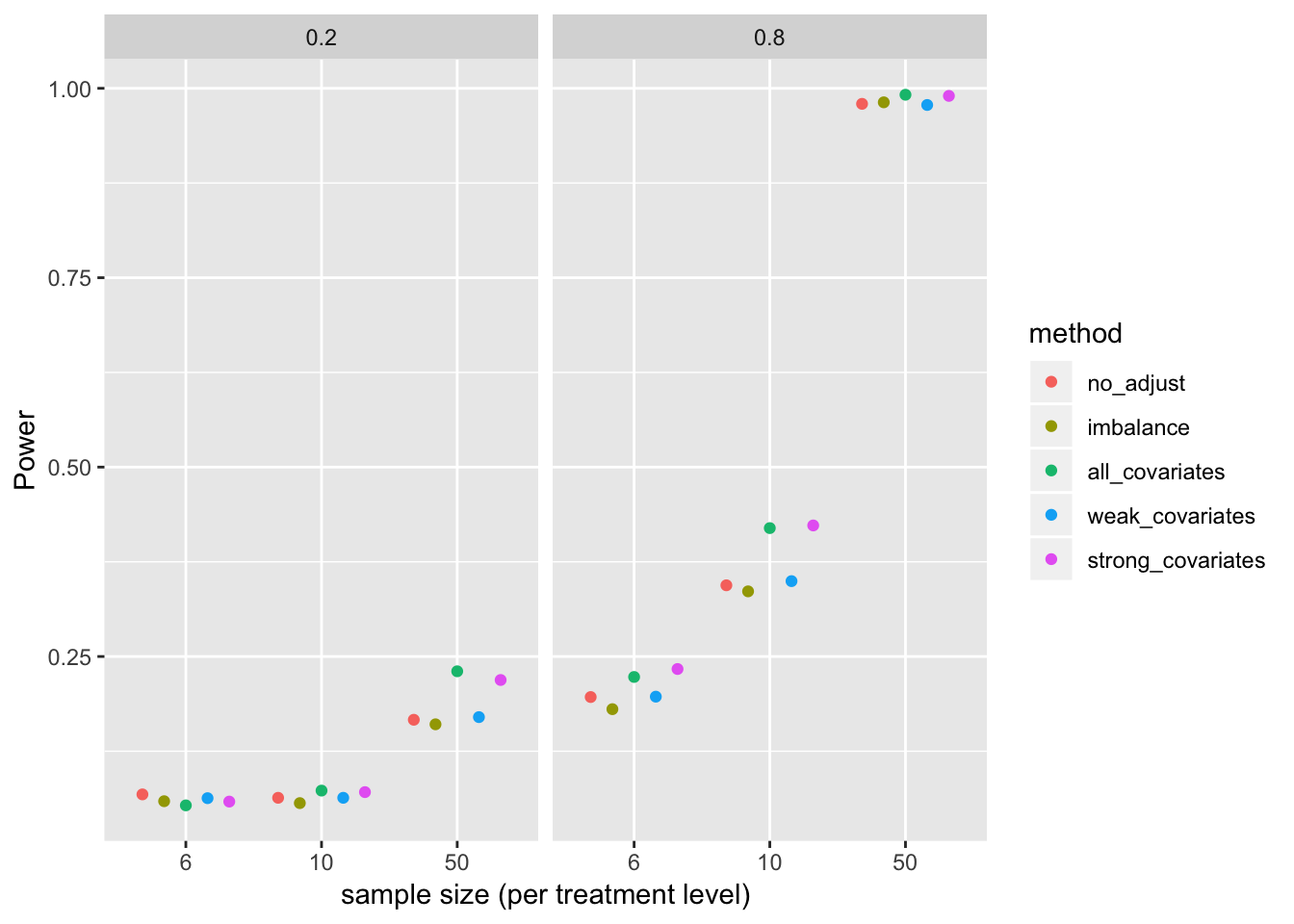
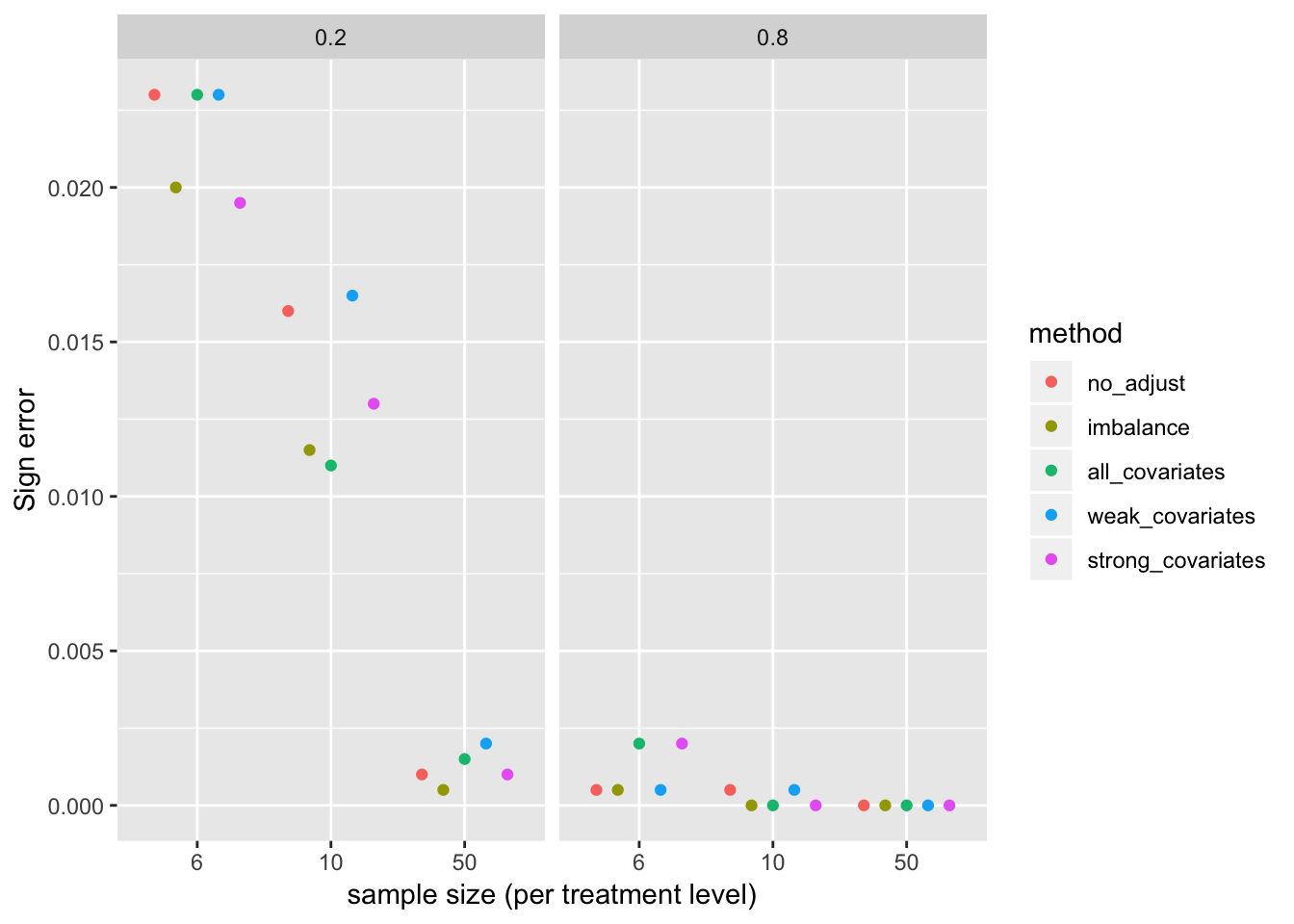
Sign error
# sign error
p_long2 <- cbind(p_long, b=b_long[, b])
sign_error <- p_long2[beta_1 > 0, .(error=sum(p < 0.1 & b < 0)/niter), by=.(method, n, beta_1)]
gg <- ggplot(data=sign_error, aes(x=factor(n), y=error, color=method)) +
geom_point(position=pd) +
xlab("sample size (per treatment level)") +
ylab("Sign error") +
facet_grid(.~beta_1) +
NULL
gg